Proteomics
Unit info
Every human cell has the same DNA and the same genes. However, the difference between cells in different tissues and organs as well as between healthy and cancer cells is that the expression of genes is different. Expression means that proteins, which are the products of the genes, are different in type, amount or modification states and these variations are the subject of Proteomics.
The Proteomics/MS TDU is a full proteomic infrastructure for several analyses ranging from simple identifications of proteins to complex analysis involving multistage separations techniques, analysis of post translational modified proteins and quantification processes, both targeted and untargeted. This includes various platforms for protein and peptide separation, and state-of-the-art mass spectrometry for MS and LC-MS/MS experiments.
The main interest of the Unit is to develop, implement and optimize proteomic based protocols along with IFOM's groups to obtain synergistic technologies to apply in cancer research. Proteomics techniques are rapidly advancing and require considerable technical expertise and expensive equipment. For this reason the aim of the IFOM Proteomics/ Mass Spectrometry Facility is to accelerate discovery in cancer proteomics by giving researchers access to cutting edge technologies in Mass Spectrometry and Proteomics.

Angela Bachi
Angela Bachi is also Principal Investigator of the Functional Proteomics program.
See her profile
Unit member
Angela CattaneoVittoria Matafora
Photogallery
 Proteomics unit 01
Proteomics unit 01 Proteomics unit 02
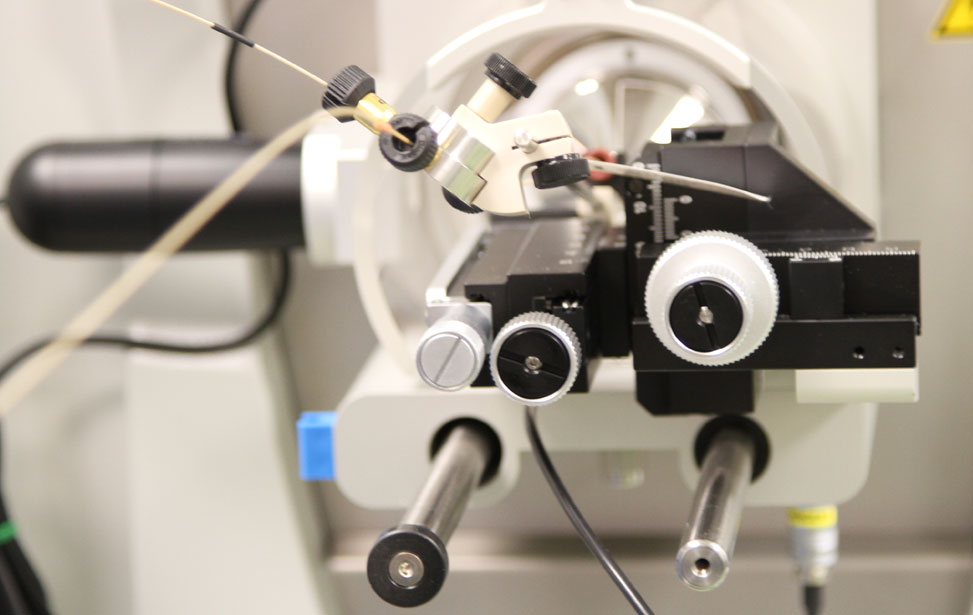
Proteomics unit 02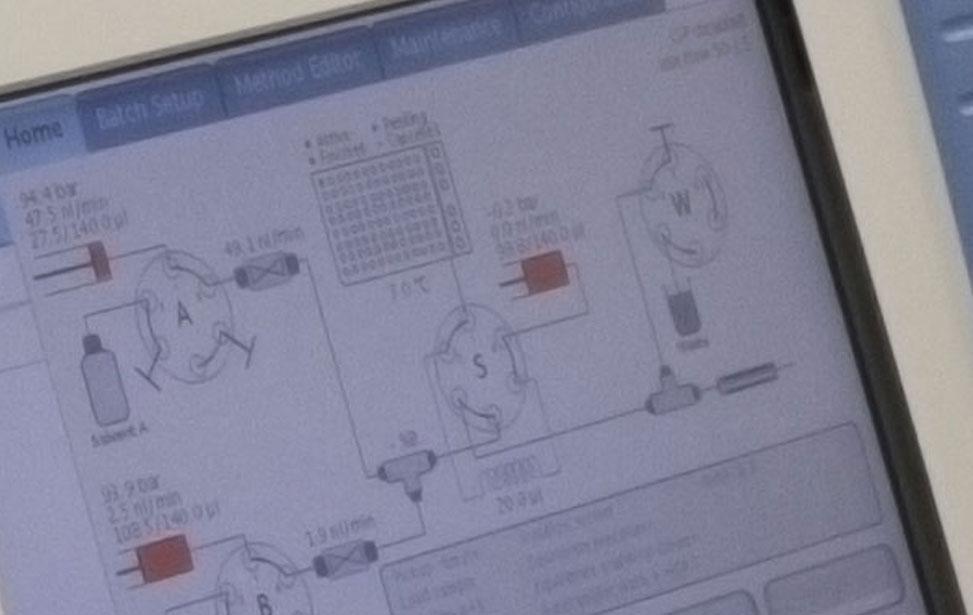 Proteomics unit 03
Proteomics unit 03 Proteomics unit 04
Proteomics unit 04 Proteomics unit 05
Proteomics unit 05 Proteomics unit 06
Proteomics unit 06Proteomics Instruments
Selected Unit Publications
- Mapping the native interaction surfaces of PREP1 with PBX1 by cross-linking mass-spectrometry and mutagenesis.
Bruckmann C, Tamburri S, De Lorenzi V, Doti N, Monti A, Mathiasen L, Cattaneo A, Ruvo M, Bachi A, Blasi F.
Sci Rep. 2020 Oct 8;10(1):16809. doi: 10.1038/s41598-020-74032-w. - IRSp53 controls plasma membrane shape and polarized transport at the nascent lumen in epithelial tubules.
Martano G, Leone M, D'Oro P, Matafora V, Cattaneo A, Masseroli M, Bachi A.
Nat Commun. 2020 Jul 14;11(1):3516. doi: 10.1038/s41467-020-17091-x. - SMfinder: Small Molecules Finder for Metabolomics and Lipidomics Analysis.
Bisi S, Marchesi S, Rizvi A, Carra D, Beznoussenko GV, Ferrara I, Deflorian G, Mironov A, Bertalot G, Pisati F, Oldani A, Cattaneo A, Saberamoli G, Pece S, Viale G, Bachi A, Tripodo C, Scita G, Disanza A.
Anal Chem. 2020 Jul 7;92(13):8874-8882. doi: 10.1021/acs.analchem.0c00585. Epub 2020 Jun 16. - ATR expands embryonic stem cell fate potential in response to replication stress.
Atashpaz S, Samadi Shams S, Gonzalez JM, Sebestyèn E, Arghavanifard N, Gnocchi A, Albers E, Minardi S, Faga G, Soffientini P, Allievi E, Cancila V, Bachi A, Fernàndez-Capetillo Ó, Tripodo C, Ferrari F, LÓpez-Contreras AJ, Costanzo V.
Elife. 2020 Mar 12;9:e54756. doi: 10.7554/eLife.54756. - ATR is essential for preservation of cell mechanics and nuclear integrity during interstitial migration.
Kidiyoor GR, Li Q, Bastianello G, Bruhn C, Giovannetti I, Mohamood A, Beznoussenko GV, Mironov A, Raab M, Piel M, Restuccia U, Matafora V, Bachi A, Barozzi S, Parazzoli D, Frittoli E, Palamidessi A, Panciera T, Piccolo S, Scita G, Maiuri P, Havas KM, Zhou ZW, Kumar A, Bartek J, Wang ZQ, Foiani M.
Nat Commun. 2020 Sep 24;11(1):4828. doi: 10.1038/s41467-020-18580-9. - Vitamin C Restricts the Emergence of Acquired Resistance to EGFR-Targeted Therapies in Colorectal Cancer.
Lorenzato A, Magrì A, Matafora V, Audrito V, Arcella P, Lazzari L, Montone M, Lamba S, Deaglio S, Siena S, Bertotti A, Trusolino L, Bachi A, Di Nicolantonio F, Bardelli A, Arena S.
Cancers (Basel). 2020 Mar 14;12(3):685. doi: 10.3390/cancers12030685. - STAGE-Diging in Proteomics.
Soffientini P, Bachi A.
Methods Mol Biol. 2017;1654:255-260. doi: 10.1007/978-1-4939-7231-9_18. - Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression.
Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V.
Nat Cell Biol. 2016 Jun;18(6):684-91. doi: 10.1038/ncb3344. - SEPN1, an endoplasmic reticulum-localized selenoprotein linked to skeletal muscle pathology, counteracts hyperoxidation by means of redox-regulating SERCA2 pump activity.
Marino M, Stoilova T, Giorgi C, Bachi A, Cattaneo A, Auricchio A, Pinton P, Zito E.
Hum Mol Genet. 2014 Dec 1. - Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming.
Maspero E, Valentini E, Mari S, Cecatiello V, Soffientini P, Pasqualato S, Polo S.
Nat Struct Mol Biol. 2013 Jun;20(6):696-701


