Quantitative Biology of Cell Division
Research project
Our lab is interested in studying molecular regulatory networks using both computational and experimental approaches.
Metaphase-to-anaphase transition
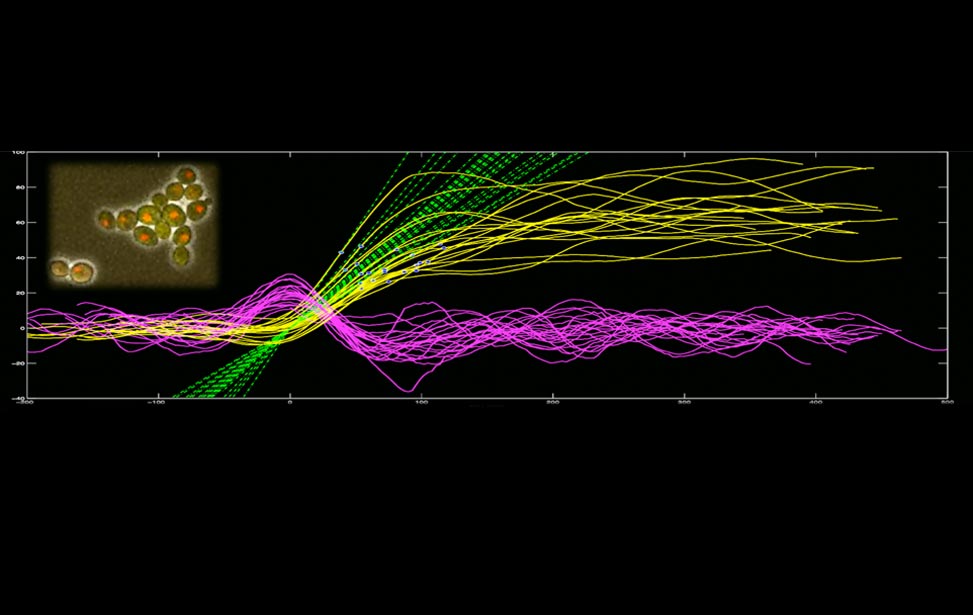
Our main interest lies in the analysis of the general principles of cell cycle control, with particular emphasis on the exit from mitosis. Normally, cells make sure that the two daughter cells born at the end of a cell cycle inherit one sister chromatid each, and thus are genetically equivalent to their mother cell. We are interested in understanding the molecular device (known as the Spindle Assembly Checkpoint or SAC) put in place by cells to avoid an unequal segregation of genetic material at the end of mitosis. The approach we follow combines computational tools and experimental approaches to isolate molecular circuits that are known to play important roles in the checkpoint. With this approach, we have recently shown that a positive feedback loop can ontribute to the establishment of the checkpoint [The influence of catalysis on mad2 activation dynamics. Simonetta M, Manzoni R, Mosca R, Mapelli M, Massimiliano L, Vink M, Novak B, Musacchio A, Ciliberto A. PLoS Biol. 2009 Jan 13;7(1):e10.], and we are further investigating how this circuit could contribute to the quick inactivation of the checkpoint before the metaphase-to-anaphase transition.
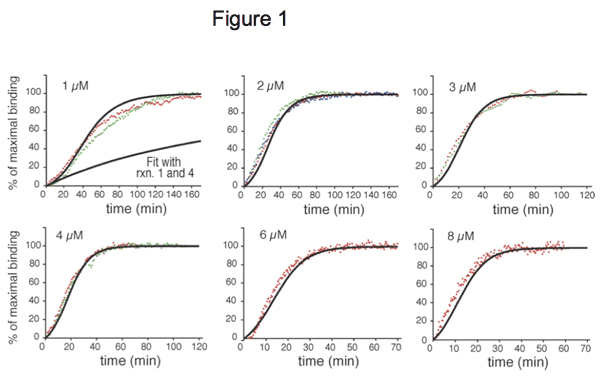
Figure 1. Experimental and simulated kinetics of Cdc20:Mad2 binding in vitro, for different doses of Mad2, from Simonetta et al. 2010
Cell cycle regulation
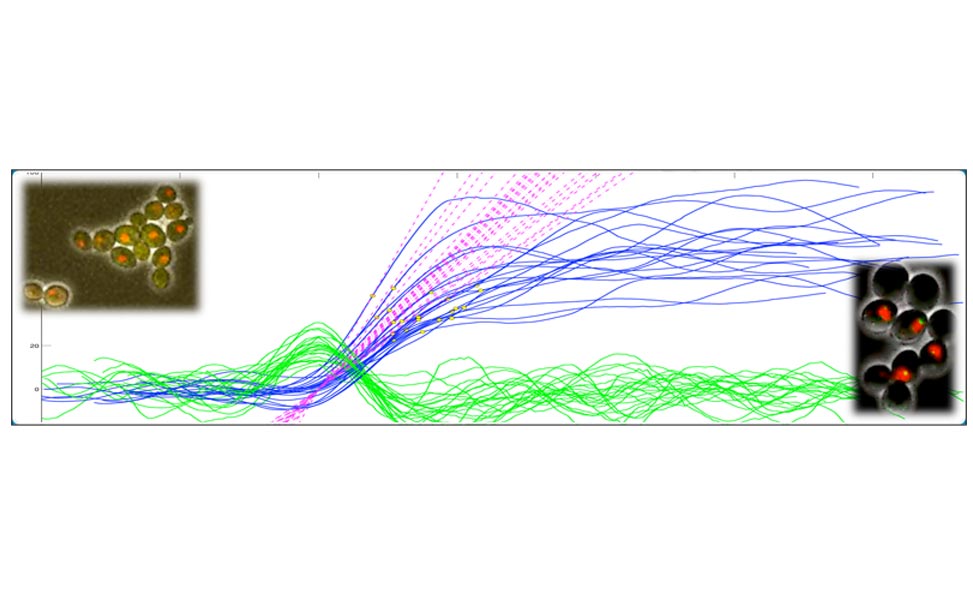
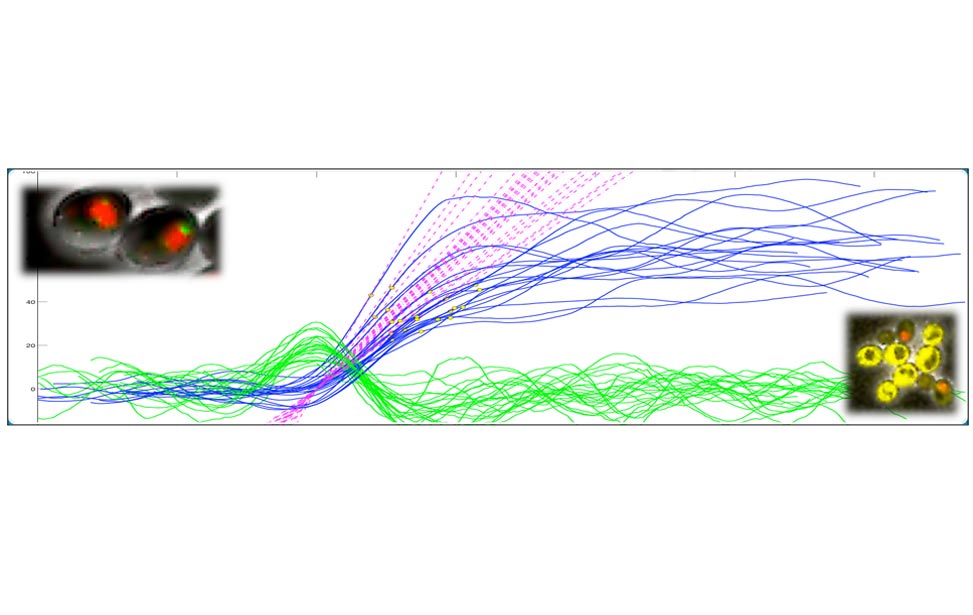
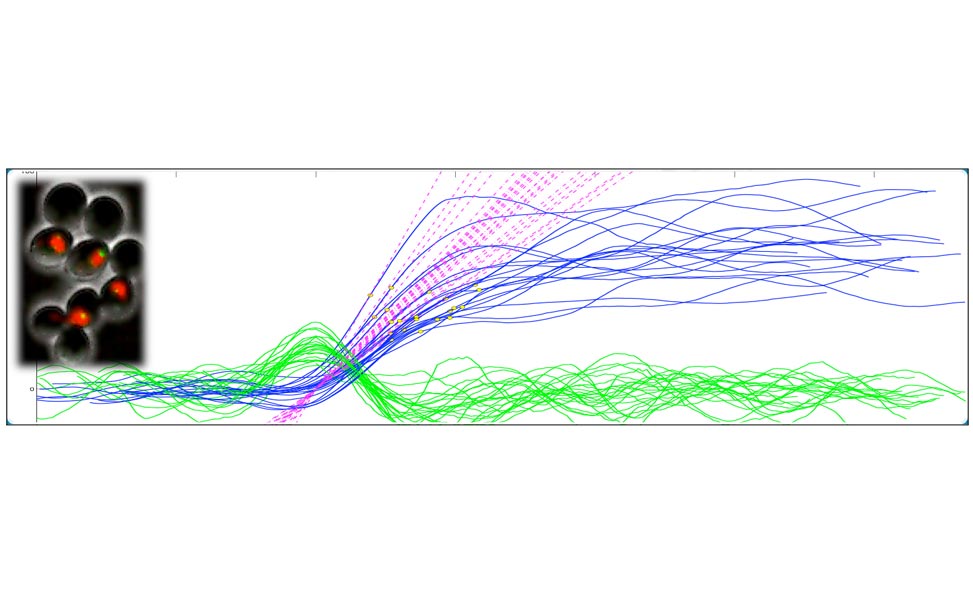
Entry and exit from mitosis are driven by the activation and inactivation of the key regulators of mitosis, the complexes formed by Cyclin Dependent Kinases (CDK) and mitotic cyclins. The cycle of CDK-Cyclin activation, sometimes called the cell-cycle engine, drives other periodical processes such as DNA replication the centrosome cycle, the release and re-sequestration of mitotic regulators (e.g., the protein phosphatase Cdc14 in yeast). We are thus interested in analyzing the mechanism whereby the cell cycle engine triggers these additional, ancillary events. As a test case, we started from Cdc14 localization, an event which we have shown to be capable of oscillatory behavior [Oscillations in Cdc14 release and sequestration reveal a circuit underlying mitotic exit. Manzoni R, Montani F, Visintin C, Caudron F, Ciliberto A, Visintin R. J Cell Biol. 2010 Jul 26;190(2):209-22.] in the presence of high and constant values of CDK-(mitotic) Cyclin levels.
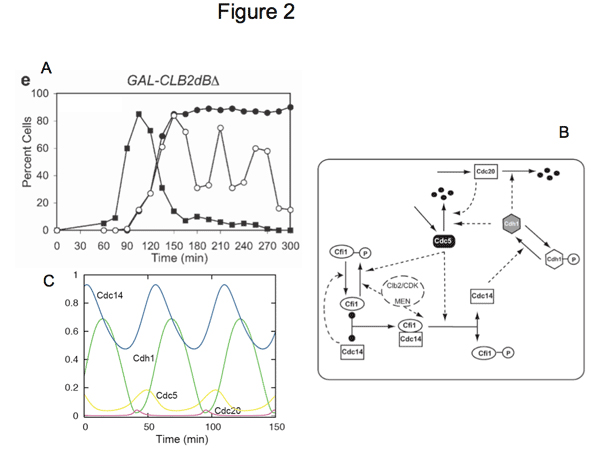
Figure 2. Oscillations of Cdc14 outside/inside the nucleolus can be observed in cells blocked in mitosis by non-degradable Clb2 (A). The molecular network that presumably contributes to the oscillatory behavior is shown in (B) and simulations of the network are shown in (C). Figures adapted from Manzoni et al. 2010.
30.01.2026